Publications
2025
-
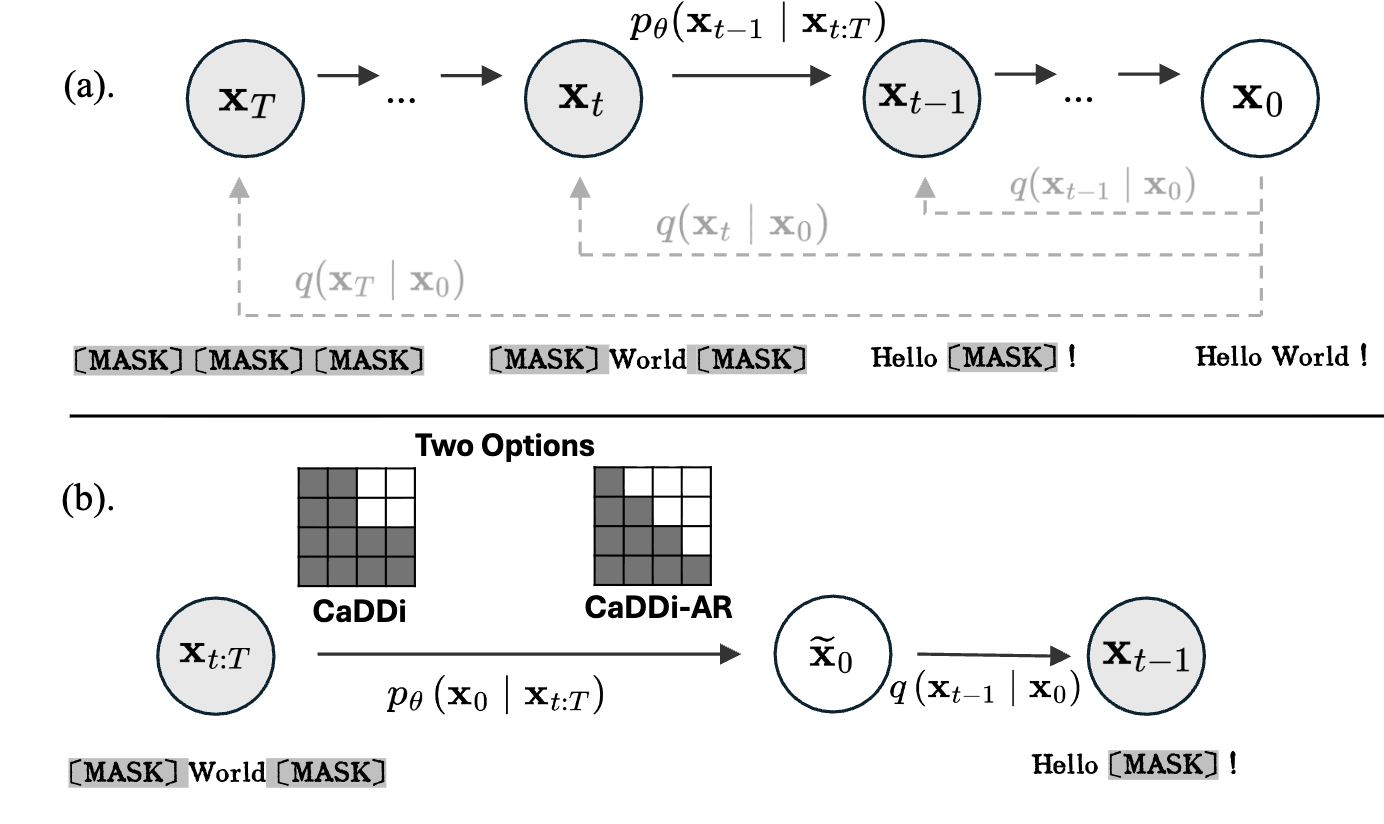 Non-Markovian Discrete Diffusion with Causal Language ModelsYangtian Zhang*, Sizhuang He*, Daniel Levine, Lawrence Zhao, David Zhang, Syed A Rizvi, Emanuele Zappala, Rex Ying, and 1 more authorAdvances in Neural Information Processing Systems, 2025
Non-Markovian Discrete Diffusion with Causal Language ModelsYangtian Zhang*, Sizhuang He*, Daniel Levine, Lawrence Zhao, David Zhang, Syed A Rizvi, Emanuele Zappala, Rex Ying, and 1 more authorAdvances in Neural Information Processing Systems, 2025Discrete diffusion models offer a flexible, controllable approach to structured sequence generation, yet they still lag behind causal language models in expressive power. A key limitation lies in their reliance on the Markovian assumption, which restricts each step to condition only on the current state, leading to potential uncorrectable error accumulation. In this paper, we introduce CaDDi, a discrete diffusion model that conditions on the entire generative trajectory, thereby lifting the Markov constraint and allowing the model to revisit and improve past states. By unifying sequential (causal) and temporal (diffusion) reasoning in a single non-Markovian transformer, CaDDi also treats standard causal language models as a special case and permits the direct reuse of pretrained LLM weights with no architectural changes. Empirically, CaDDi outperforms state-of-the-art discrete diffusion baselines on natural-language benchmarks, substantially narrowing the remaining gap to large autoregressive transformers.
@article{zhang2025caddi, title = {Non-Markovian Discrete Diffusion with Causal Language Models}, author = {Zhang, Yangtian and He, Sizhuang and Levine, Daniel and Zhao, Lawrence and Zhang, David and Rizvi, Syed A and Zappala, Emanuele and Ying, Rex and van Dijk, David}, journal = {Advances in Neural Information Processing Systems}, year = {2025}, } -
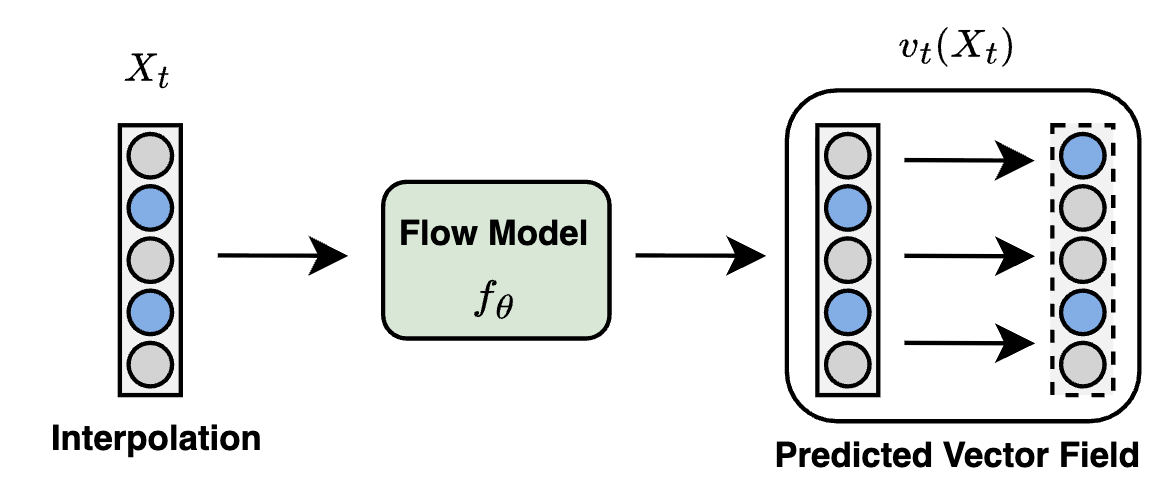 Flow Matching for Collaborative FilteringChengkai Liu*, Yangtian Zhang*, Jianling Wang, Rex Ying, and James CaverleeACM SIGKDD Conference on Knowledge Discovery and Data Mining, 2025
Flow Matching for Collaborative FilteringChengkai Liu*, Yangtian Zhang*, Jianling Wang, Rex Ying, and James CaverleeACM SIGKDD Conference on Knowledge Discovery and Data Mining, 2025Generative models have shown great promise in collaborative filtering by capturing the underlying distribution of user interests and preferences. However, existing approaches struggle with inaccurate posterior approximations and misalignment with the discrete nature of recommendation data, limiting their expressiveness and real-world performance. To address these limitations, we propose FlowCF, a novel flow-based recommendation system leveraging flow matching for collaborative filtering. We tailor flow matching to the unique challenges in recommendation through two key innovations: (1) a behavior-guided prior that aligns with user behavior patterns to handle the sparse and heterogeneous user-item interactions, and (2) a discrete flow framework to preserve the binary nature of implicit feedback while maintaining the benefits of flow matching, such as stable training and efficient inference. Extensive experiments demonstrate that FlowCF achieves state-of-the-art recommendation accuracy across various datasets with the fastest inference speed, making it a compelling approach for real-world recommender systems. The code is available at https://github.com/chengkai-liu/FlowCF.
@article{liu2025flow, title = {Flow Matching for Collaborative Filtering}, author = {Liu, Chengkai and Zhang, Yangtian and Wang, Jianling and Ying, Rex and Caverlee, James}, journal = {ACM SIGKDD Conference on Knowledge Discovery and Data Mining}, year = {2025}, }
2024
-
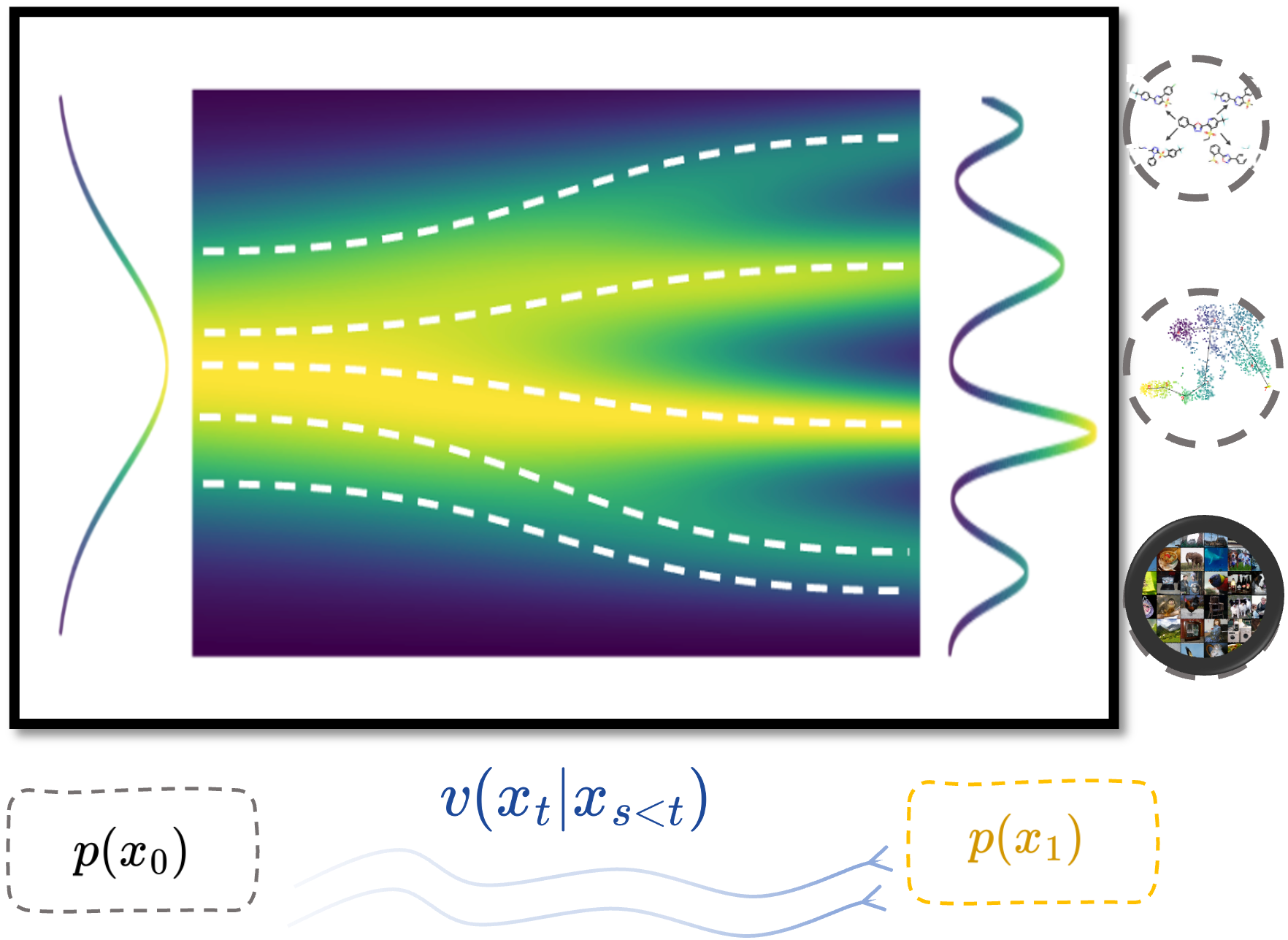 CaLMFlow: Volterra Flow Matching using Causal Language ModelsSizhuang He, Daniel Levine, Ivan Vrkic, Marco Francesco Bressana, David Zhang, Syed Asad Rizvi, Yangtian Zhang, Emanuele Zappala, and 1 more authorarXiv preprint arXiv:2410.05292, 2024
CaLMFlow: Volterra Flow Matching using Causal Language ModelsSizhuang He, Daniel Levine, Ivan Vrkic, Marco Francesco Bressana, David Zhang, Syed Asad Rizvi, Yangtian Zhang, Emanuele Zappala, and 1 more authorarXiv preprint arXiv:2410.05292, 2024We present CaLMFlow, a novel framework for Volterra flow matching using causal language models. Our approach leverages the causal nature of language models to efficiently compute the conditional score function required for training score-based models. By incorporating the score matching objective into the training process of language models, we enable the generation of high-quality samples from the model. We demonstrate the effectiveness of our method on a diverse set of tasks, including image generation, text-to-image synthesis, and molecular dynamics simulation. Our experiments show that CaLMFlow can generate realistic samples comparable to state-of-the-art score-based models while being significantly more efficient in both training and inference. Code is available at https://github.com/calmflow/calmflow.
@article{he2024calmflow, title = {CaLMFlow: Volterra Flow Matching using Causal Language Models}, author = {He, Sizhuang and Levine, Daniel and Vrkic, Ivan and Bressana, Marco Francesco and Zhang, David and Rizvi, Syed Asad and Zhang, Yangtian and Zappala, Emanuele and van Dijk, David}, journal = {arXiv preprint arXiv:2410.05292}, year = {2024}, }
2023
-
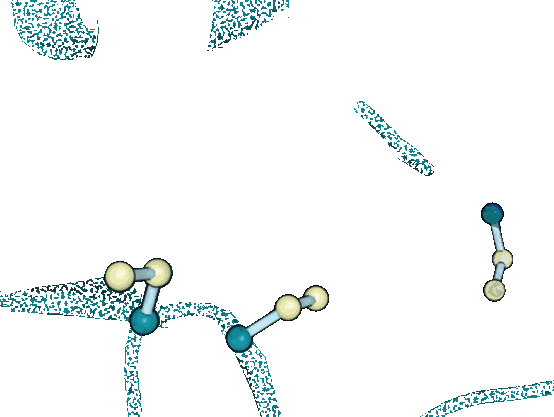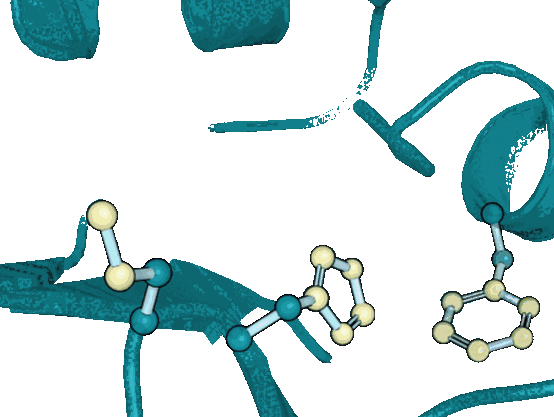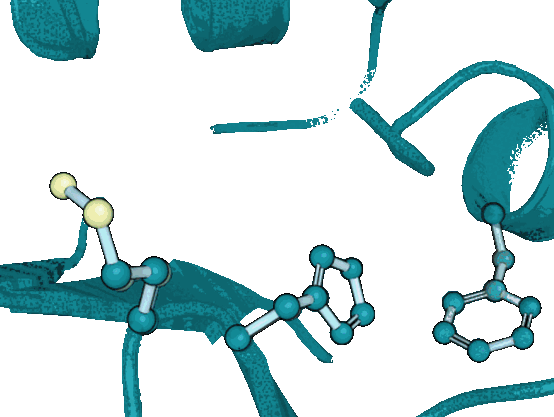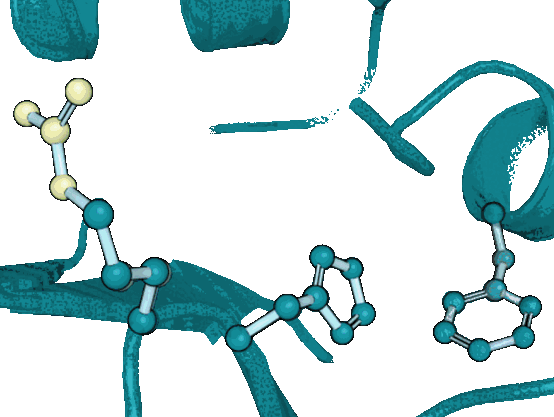 DiffPack: A Torsional Diffusion Model for Autoregressive Protein Side-Chain PackingYangtian Zhang*, Zuobai Zhang*, Bozitao Zhong, Sanchit Misra, and Jian TangAdvances in Neural Information Processing Systems, 2023
DiffPack: A Torsional Diffusion Model for Autoregressive Protein Side-Chain PackingYangtian Zhang*, Zuobai Zhang*, Bozitao Zhong, Sanchit Misra, and Jian TangAdvances in Neural Information Processing Systems, 2023Proteins play a critical role in carrying out biological functions, and their 3D structures are essential in determining their functions. Accurately predicting the conformation of protein side-chains given their backbones is important for applications in protein structure prediction, design and protein-protein interactions. Traditional methods are computationally intensive and have limited accuracy, while existing machine learning methods treat the problem as a regression task and overlook the restrictions imposed by the constant covalent bond lengths and angles. In this work, we present DiffPack, a torsional diffusion model that learns the joint distribution of side-chain torsional angles, the only degrees of freedom in side-chain packing, by diffusing and denoising on the torsional space. To avoid issues arising from simultaneous perturbation of all four torsional angles, we propose autoregressively generating the four torsional angles from \chi_1 to \chi_4 and training diffusion models for each torsional angle. We evaluate the method on several benchmarks for protein side-chain packing and show that our method achieves improvements of 11.9% and 13.5% in angle accuracy on CASP13 and CASP14, respectively, with a significantly smaller model size (60x fewer parameters). Additionally, we show the effectiveness of our method in enhancing side-chain predictions in the AlphaFold2 model. Code will be available upon the accept.
@article{zhan2023diffpack, title = {DiffPack: A Torsional Diffusion Model for Autoregressive Protein Side-Chain Packing}, author = {Zhang, Yangtian and Zhang, Zuobai and Zhong, Bozitao and Misra, Sanchit and Tang, Jian}, journal = {Advances in Neural Information Processing Systems}, year = {2023}, } -
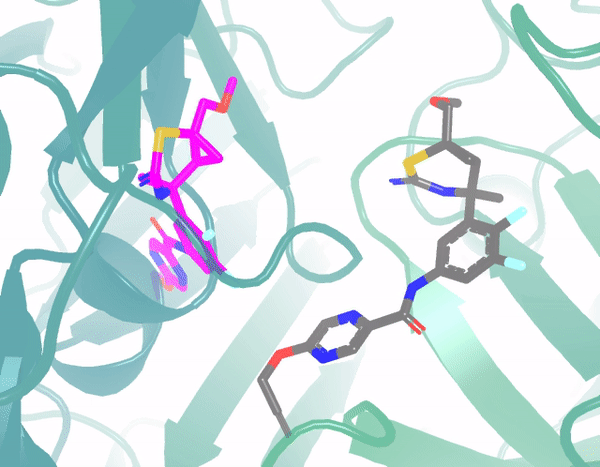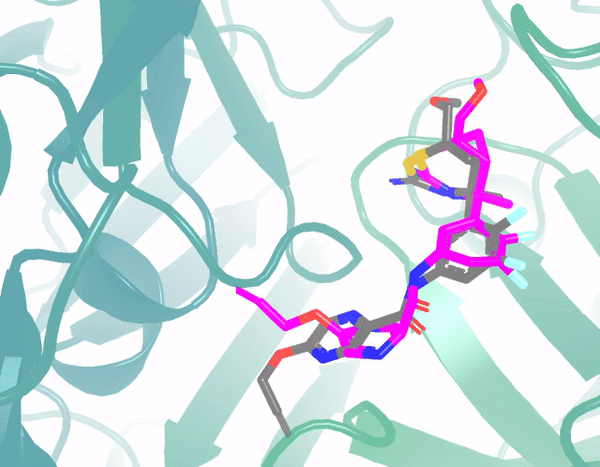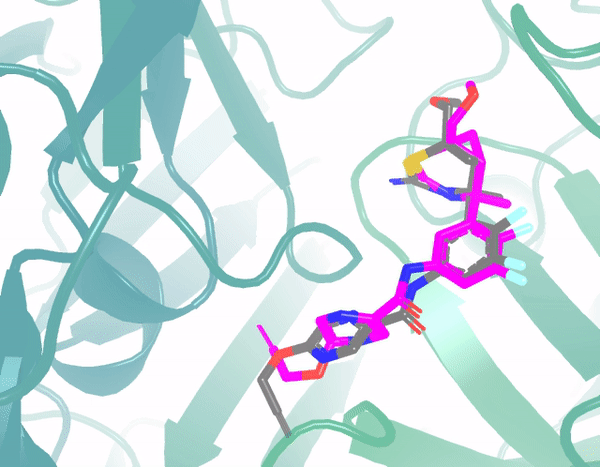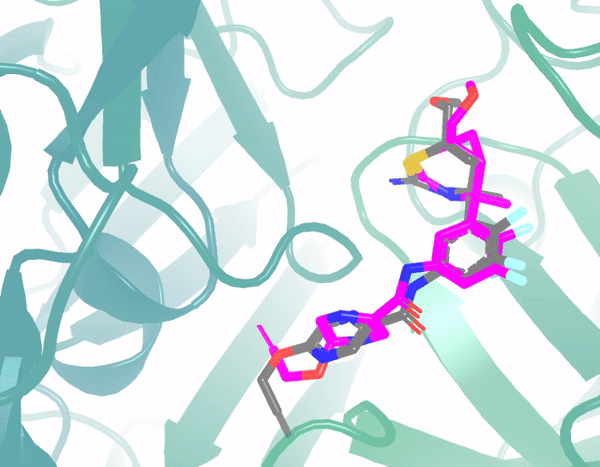 E3Bind: An End-to-End Equivariant Network for Protein-Ligand DockingYangtian Zhang*, Huiyu Cai*, Chence Shi, Bozitao Zhong, and Jian TangProceedings of the International Conference on Learning Representations (ICLR), 2023
E3Bind: An End-to-End Equivariant Network for Protein-Ligand DockingYangtian Zhang*, Huiyu Cai*, Chence Shi, Bozitao Zhong, and Jian TangProceedings of the International Conference on Learning Representations (ICLR), 2023In silico prediction of the ligand binding pose to a given protein target is a crucial but challenging task in drug discovery. This work focuses on blind flexible selfdocking, where we aim to predict the positions, orientations and conformations of docked molecules. Traditional physics-based methods usually suffer from inaccurate scoring functions and high inference cost. Recently, data-driven methods based on deep learning techniques are attracting growing interest thanks to their efficiency during inference and promising performance. These methods usually either adopt a two-stage approach by first predicting the distances between proteins and ligands and then generating the final coordinates based on the predicted distances, or directly predicting the global roto-translation of ligands. In this paper, we take a different route. Inspired by the resounding success of AlphaFold2 for protein structure prediction, we propose E3Bind, an end-to-end equivariant network that iteratively updates the ligand pose. E3Bind models the protein-ligand interaction through careful consideration of the geometric constraints in docking and the local context of the binding site. Experiments on standard benchmark datasets demonstrate the superior performance of our end-to-end trainable model compared to traditional and recently-proposed deep learning methods.
@article{zhang2022e3bind, title = {E3Bind: An End-to-End Equivariant Network for Protein-Ligand Docking}, author = {Zhang, Yangtian and Cai, Huiyu and Shi, Chence and Zhong, Bozitao and Tang, Jian}, journal = {Proceedings of the International Conference on Learning Representations (ICLR)}, year = {2023}, }
2022
-
 PEER: A Comprehensive and Multi-task Benchmark for Protein Sequence UnderstandingMinghao Xu, Zuobai Zhang, Jiarui Lu, Zhaocheng Zhu, Yangtian Zhang, Ma Chang, Runcheng Liu, and Jian TangAdvances in Neural Information Processing Systems (Datasets and Benchmarks Track), 2022
PEER: A Comprehensive and Multi-task Benchmark for Protein Sequence UnderstandingMinghao Xu, Zuobai Zhang, Jiarui Lu, Zhaocheng Zhu, Yangtian Zhang, Ma Chang, Runcheng Liu, and Jian TangAdvances in Neural Information Processing Systems (Datasets and Benchmarks Track), 2022@article{xu2022peer, title = {PEER: A Comprehensive and Multi-task Benchmark for Protein Sequence Understanding}, author = {Xu, Minghao and Zhang, Zuobai and Lu, Jiarui and Zhu, Zhaocheng and Zhang, Yangtian and Chang, Ma and Liu, Runcheng and Tang, Jian}, journal = {Advances in Neural Information Processing Systems (Datasets and Benchmarks Track)}, volume = {35}, pages = {35156--35173}, year = {2022}, } -
 TorchDrug: A Powerful and Flexible Machine Learning Platform for Drug DiscoveryZhaocheng Zhu, Chence Shi, Zuobai Zhang, Shengchao Liu, Minghao Xu, Xinyu Yuan, Yangtian Zhang, Junkun Chen, and 3 more authorsPreprint, 2022
TorchDrug: A Powerful and Flexible Machine Learning Platform for Drug DiscoveryZhaocheng Zhu, Chence Shi, Zuobai Zhang, Shengchao Liu, Minghao Xu, Xinyu Yuan, Yangtian Zhang, Junkun Chen, and 3 more authorsPreprint, 2022@article{zhu2022torchdrug, title = {TorchDrug: A Powerful and Flexible Machine Learning Platform for Drug Discovery}, author = {Zhu, Zhaocheng and Shi, Chence and Zhang, Zuobai and Liu, Shengchao and Xu, Minghao and Yuan, Xinyu and Zhang, Yangtian and Chen, Junkun and Cai, Huiyu and Lu, Jiarui and others}, journal = {Preprint}, year = {2022}, } -
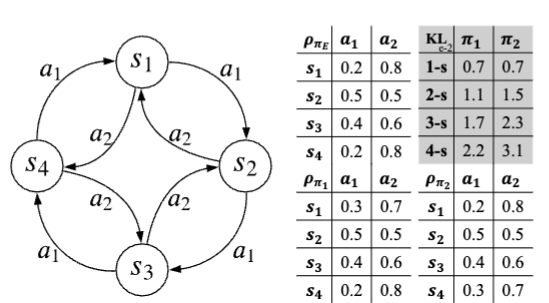 Imitation Learning via Multi-Step Occupancy Measure MatchingMinghuan Liu, Hangyu Wang, Yangtian Zhang, Minkai Xu, Zhengbang Zhu, and Weinan ZhangPreprint, 2022
Imitation Learning via Multi-Step Occupancy Measure MatchingMinghuan Liu, Hangyu Wang, Yangtian Zhang, Minkai Xu, Zhengbang Zhu, and Weinan ZhangPreprint, 2022Imitation learning (IL) aims to learn a policy from expert demonstrations without reward signals. Previous methods such as behavior cloning (BC) work by learning one-step predictions, but seriously suffer from the compounding error problem; recent generative adversarial solution, though alleviates such problems in a discrepancy minimization view, is still limited in only matching single-step state-action distributions instead of long-term trajectories. To address the long-range effect, in this paper, we explore the potential to boost the performance of IL by regularizing the multi-step discrepancies. We first propose the multi-step occupancy measure matching formulation, where we extend the idea of matching single state-action pairs to sequences of multiple steps. Interestingly, theoretical analysis of the proposed multi-step algorithm reveals a trade-off between the rollout discrepancy and the sampling complexity, making it non-trivial to select an appropriate step length T for the practical implementation. Inspired by the recent progress of integrating multi-armed bandits in curriculum learning, we further propose an automated curriculum multi-step occupancy measure matching algorithm named AutoGAIL, which automatically selects the appropriate step length during the training procedure. Compared with various multi-step GAIL baselines, AutoGAIL consistently achieves superior performance with satisfactory learning efficiency given different amount of demonstrations.
@article{liuautogail, title = {Imitation Learning via Multi-Step Occupancy Measure Matching}, author = {Liu, Minghuan and Wang, Hangyu and Zhang, Yangtian and Xu, Minkai and Zhu, Zhengbang and Zhang, Weinan}, journal = {Preprint}, year = {2022}, }